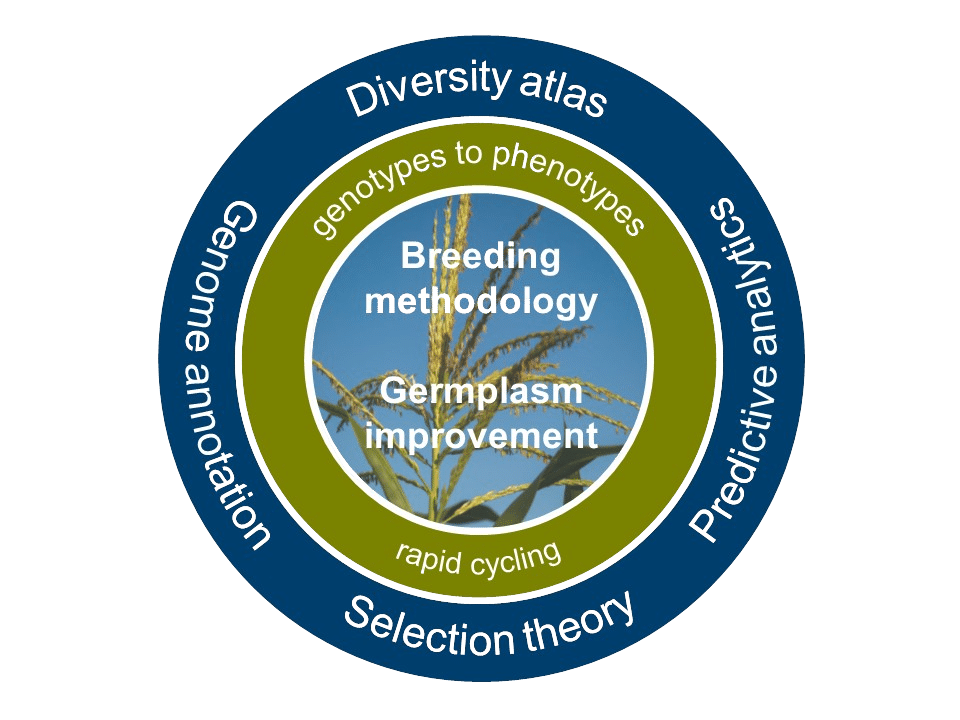
The project MAZE develops solutions to access native diversity in a targeted way with the goal to improve quantitative traits relevant for crop production. Resolving structure-function relationships at the nucleotide and amino acid level is a key step in a biology-inspired breeding strategy. Building on the achievements of previous project phases, we aim to molecularly characterize genomic regions with a large effect on early development, root and drought-related traits. The goal is to understand the function of candidate genes in these regions and to search for additional alleles in the available maize gene pool.
In addition to functional knowledge on individual genes population improvement is required to broaden the genetic base of elite germplasm for quantitative traits. Genome-based methods open new avenues for decreasing cycle length and upscaling the number of selection candidates. During the course of the MAZE project, a genome-based rapid cycling experiment was initiated. The resulting experimental data will provide a comprehensive assessment of the potential of rapid cycling selection in landraces.
MAZE is funded by the German Federal Ministry of Education and Research (BMBF) within the scope of the funding initiative “Plant Breeding Research for the Bioeconomy” (Funding ID Phase 1: 031B0195, Phase 2: 031B0882, Phase 3: 031B1301).
Technical University of Munich
Plant Breeding
plantbreeding.wzw@tum.de
© 2023 europeanmaize.net
