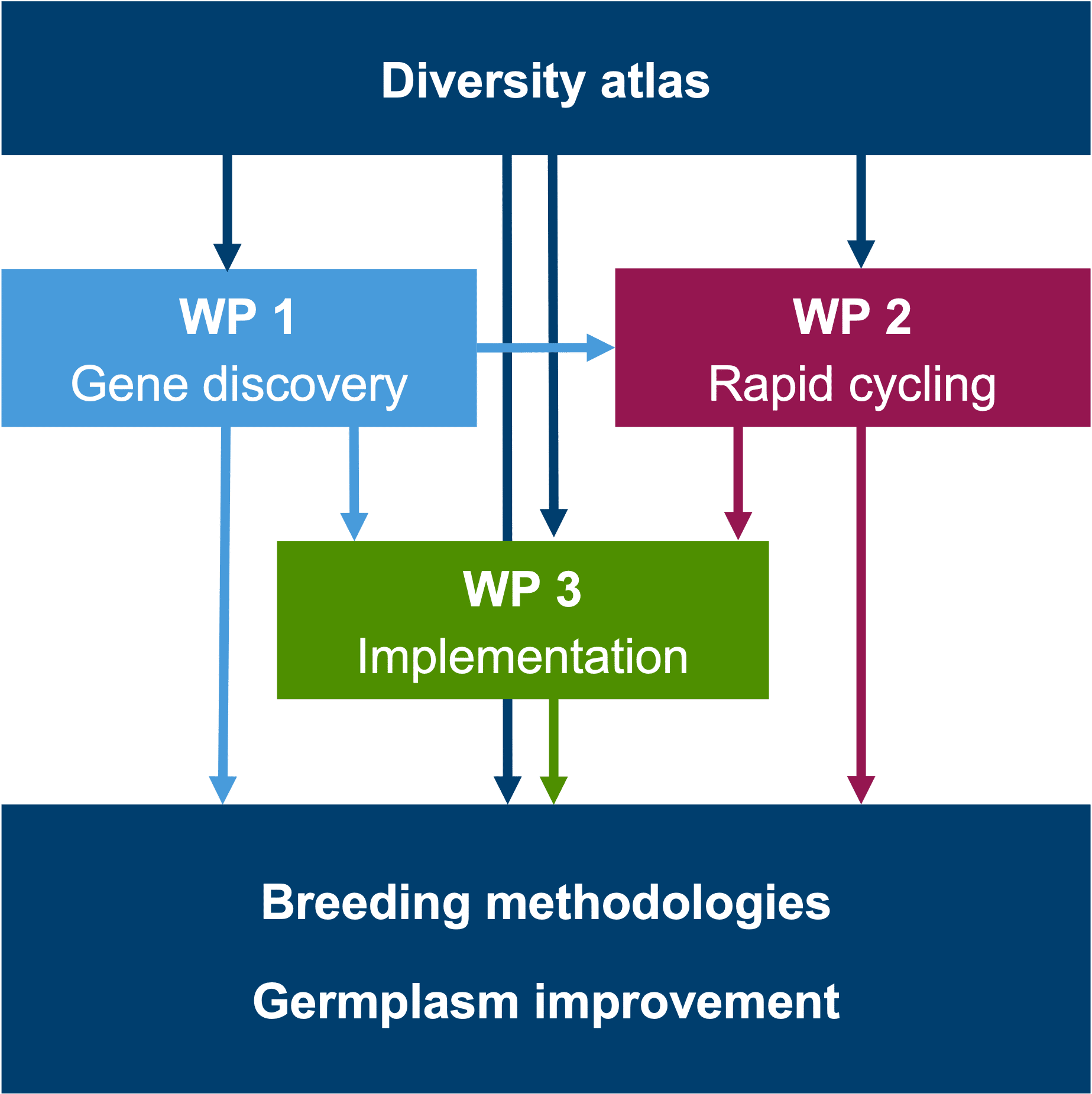
In the third phase of MAZE, the project will advance knowledge, materials and methods developed in previous phases with a strong focus on efficient, knowledge-based variety development. Material, data and results generated in MAZE allow direct implementation in breeding programs. Contributions from individual partners are highly connected within and across work packages. Partners experienced in forward and reverse genetics, in functional analyses of genes, mutant analyses, high-throughput phenotyping, maize genome structure and genome annotation will work together on gene discovery and haplotype diversity in WP1. Partners with a strong background in selection theory, quantitative and evolutionary genetics, computational methods and breeding will work together on optimization of genome-based rapid cycling selection in WP2. Results of WP1 and WP2 will flow directly into an implementation project in WP3. Results on individual genes and their allelic series will be applied through marker-assisted selection in WP3 while results from WP2 will shape the selection and bridging process when introducing material from the diversity atlas into elite germplasm.
Technical University of Munich
Plant Breeding
plantbreeding.wzw@tum.de
© 2023 europeanmaize.net
